Welcome to Iris Biotech
For better service please confirm your country and language we detected.

For better service please confirm your country and language we detected.

Thank you very much for your interest in our products. All prices listed on our website are ex-works, Germany, and may attract customs duties when imported.
You may/will be contacted by the shipping company for additional documentation that may be required by the US Customs for clearance.
We offer you the convenience of buying through a local partner, Peptide Solutions LLC who can import the shipment as well as prepay the customs duties and brokerage on your behalf and provide the convenience of a domestic sale.
Continue to Iris Biotech GmbHSend request to US distributorPublished on 27.04.2016
Rhodamines are a group of fluorescent dyes that belong to the family of fluorone dyes. Rhodamine dyes have a 300-fold higher sensitivity than analogous coumarin derivatives and generate less background noise. Due to the red-shifted excitation and emission wavelengths, Rhodamine 110 interferes less with components of color-based assays compared to coumarin derivatives such as AMC (AMC: Ex 380 nm / Em 460 nm vs. Rh110: Ex 492 nm / Em 529 nm).
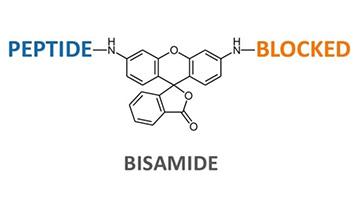
Many Rhodamine 110 protease substrates are symmetric. In symmetric substrates, the dye is bound via both amino functionalities to short identical amino acid sequences (bisamide). Cleavage of the first peptide bond to Rhodamine 110 significantly increases the compound’s fluorescence by approx. 3500, which is one of the advantages of these substrates. After the first proteolytic cleavage, symmetric Rhodamine substrates undergo a second proteolytic cleavage step, which complicates the determination of protease kinetics.
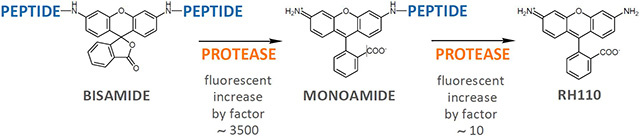
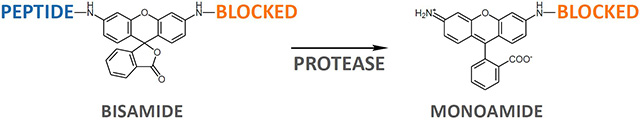
In our new asymmetric substrates, one peptide sequence is replaced by a blocking group such as D-Proline, so that only one proteolytic cleavage per substrate molecule occurs. This results in significantly simpler kinetics upon analysis, while retaining all the advantages listed above for the symmetric substrates.
References