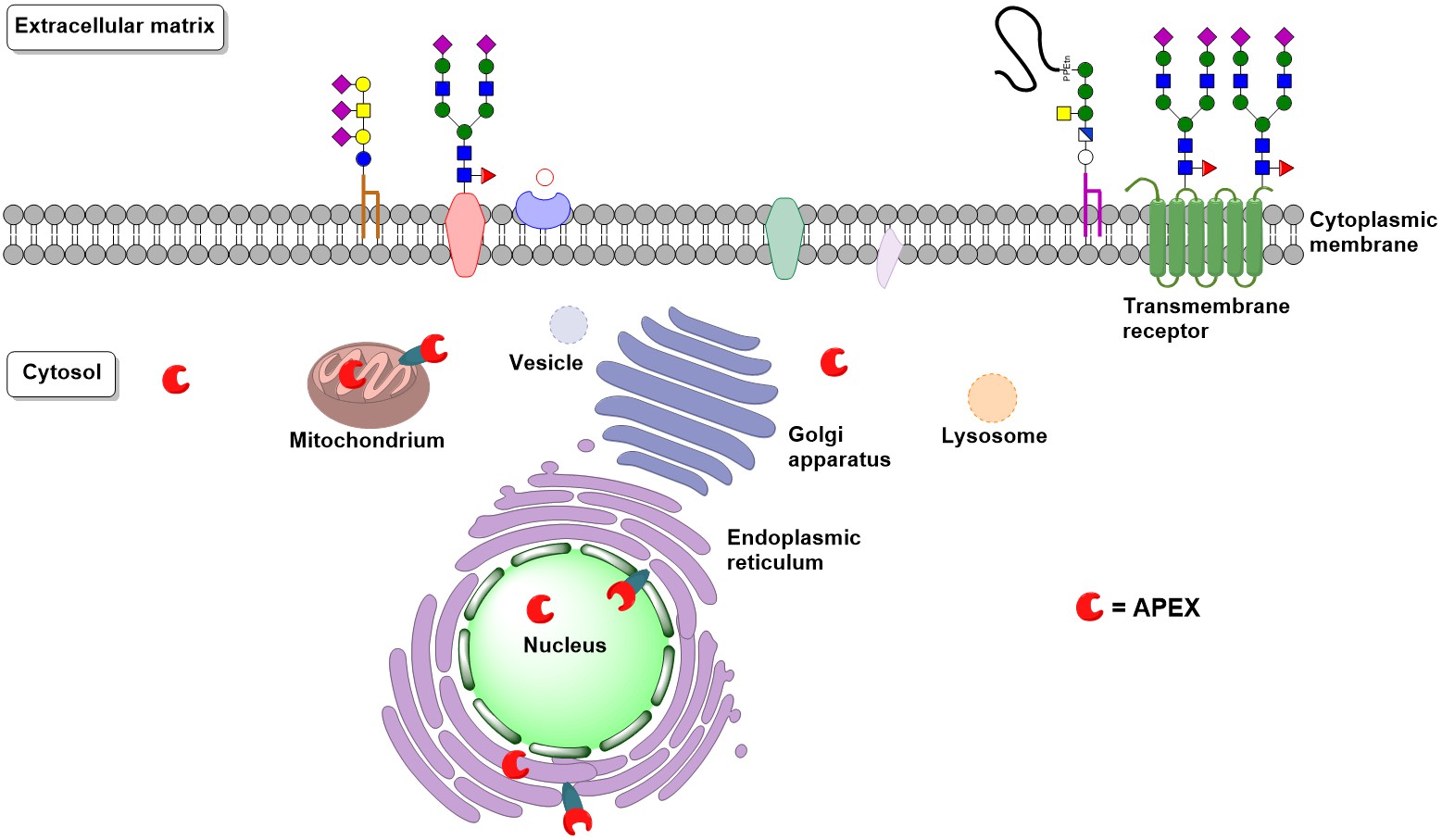
Proximity labeling of proteins with biotin-phenol (biotin-tyramide) and an engineered peroxidase enzyme (e.g. APEX = Engineered Ascorbate Peroxidase) is a pivotal tool for molecular biology.
So far, mainly phenolic compounds have been used as substrates for APEX enzymes. The enzymatic reaction yields phenoxyl radicals that predominantly react with electron-rich amino acid side chains. Consequently, phenolic compounds conjugated to biotin are not ideal for the biotinylation of nucleic acids. In order to facilitate the efficient labeling of nucleic acids, arylamine-biotin conjugates were developed (Zhou
et al., Angew. Chem. Int. Ed. 2019). These novel probes show a significantly higher reactivity towards nucleic acids compared to biotin-phenol.

APEX2 can be genetically coded to the cellular region of interest, where Biotin-Aniline allows for a biotinylation of RNA with high spatiotemporal resolution.
In combination with the peroxidase APEX2, these probes allow for the mapping of the subcellular transcriptome by proximity-dependent labeling in living cells, and may aid in elucidating the subcellular organization of biomolecules. When combined with RNA sequencing (APEX-seq), this labeling method provides complete sequence information for various classes of RNA with a high spatiotemporal resolution (Fazal & Han
et al., Cell 2019).
→ Order our catalogue Biotinylation Reagents, or download the E-Book, to get a comprehensive overview of our compounds for biotinylation.
References:
- Atlas of Subcellular RNA Localization Revealed by APEX-Seq; F. M. Fazal, S. Han, K. R. Parker, P. Kaewsapsak, J. Xu, A. N. Boettiger, H. Y. Chang and A. Y. Ting; Cell 2019; 178: 473-490 e26. doi:10.1016/j.cell.2019.05.027
- Expanding APEX2 Substrates for Proximity-Dependent Labeling of Nucleic Acids and Proteins in Living Cells; Y. Zhou, G. Wang, P. Wang, Z. Li, T. Yue, J. Wang and P. Zou; Angewandte Chemie (International ed. in English) 2019; 58: 11763-11767. doi:10.1002/anie.201905949
- Directed evolution of APEX2 for electron microscopy and proximity labeling; S. S. Lam, J. D. Martell, K. J. Kamer, T. J. Deerinck, M. H. Ellisman, V. K. Mootha and A. Y. Ting; Nat Methods 2015; 12 : 51-4. doi:10.1038/nmeth.3179